When Steven Benner set out to re-engineer genetic molecules, he didn’t think much of DNA. “The first thing you realize is that it is a stupid design,” says Benner, a biological chemist at the Foundation for Applied Molecular Evolution in Gainesville, Florida.
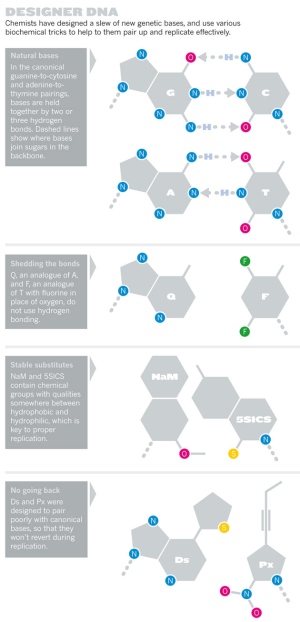
Take DNA’s backbone, which contains repeating, negatively charged phosphate groups. Because negative charges repel each other, this feature should make it harder for two DNA strands to stick together in a double helix. Then there are the two types of base-pairing: adenine (A) to thymine (T) and cytosine (C) to guanine (G). Both pairs are held together by hydrogen bonds, but those bonds are weak and easily broken up by water, something that the cell is full of. “You’re trusting your valuable genetic inheritance that you’re sending on to your children to hydrogen bonds in water?” says Benner. “If you were a chemist setting out to design this thing, you wouldn’t do it this way at all.”
Life may have had good reasons for settling on this structure, but that hasn’t stopped Benner and others from trying to change it. Over the past few decades, they have tinkered with DNA’s basic building blocks and developed a menagerie of exotic letters beyond A, T, C and G that can partner up and be copied in similar ways. But the work has presented “one goddamn problem after another”, says Benner. So far, only a few of these unnatural base pairs can be inserted into DNA consecutively, and cells are still not able to fully adopt the foreign biochemistry.
The re-engineering of DNA, and its cousin RNA, has practical goals. Artificial base pairs are already used to detect viruses and may find other uses in medicine. But scientists are also driven by the sheer novelty of it all. Eventually, they hope to develop organisms with an expanded genetic alphabet that can store more information, or perhaps ones driven by a genome with no natural letters at all. In creating these life forms, researchers could learn more about the fundamental constraints on the structure of genetic molecules and determine whether the natural bases are necessary for life or simply one solution of many. “Earth has done it a certain way in its biology,” says Gerald Joyce, a nucleic-acid biochemist at the Scripps Research Institute in La Jolla, California. “But in principle there are other ways to achieve those ends.”
Benner first became interested in those other ways as a graduate student in the 1970s. Chemists had synthesized everything from peptides to poisons, and some were trying to build molecules that could accomplish the same functions as natural enzymes or antibodies with different chemical structures. But DNA was largely ignored, he recalls. “Chemists were looking at every other class of molecule from a design perspective except the one at the centre of biology,” says Benner.
In 1986, Benner started a lab at the Swiss Federal Institute of Technology in Zurich and began to rebuild DNA’s backbone. He quickly realized that what seemed like a flaw might be a feature. When he and his team replaced the backbone’s negatively charged phosphates with neutral chemical groups1, they found that any strand longer than about a dozen units folded up on itself — probably because repelling charges were needed to keep the molecule stretched out.
The bases proved more amenable to tinkering. Benner set out to create base pairs that are similar to nature’s, but with rearranged hydrogen bonding units.
His team tested two new pairs: iso-C and iso-G (ref. 2) and ? and xanthosine3. It showed that polymerase enzymes — which copy DNA or transcribe it into RNA — could read DNA containing the unnatural bases and insert the complementary partners into a growing DNA or RNA strand. Ribosomes, the cellular machines that ‘translate’ RNA into protein, could also read an RNA snippet containing iso-C and use it to add an unnatural amino acid to a growing protein4. “The base pairing, which is at the centre of genetics, turned out to be for us the most malleable part of the molecule,” says Benner. The researchers did encounter a problem, however. Because its hydrogen atoms tend to move around, iso-G often morphed into a different form and paired with T instead of iso-C.
Unnatural bonds
Eric Kool, a chemist now at Stanford University in California, wondered whether his team could develop unnatural bases with fixed hydrogen-bonding arrangements. He and his colleagues made a base similar to the natural base T, but with fluorine in place of the oxygen atoms (see ‘Designer DNA’), among other differences5. The structure of the new base, called difluorotoluene (designated F), mimicked T’s shape almost exactly but discouraged hydrogen from jumping.
The team soon discovered that F was actually terrible at hydrogen bonding5, but polymerases still treated it like a T: during DNA copying, they faithfully inserted A opposite F (ref. 6) and vice versa7. The work suggested that as long as the base had the right shape, a polymerase could slot it in correctly. “If the key fits, it works,” says Kool.
Other scientists were dubious. “I got outraged e-mails from people saying, ‘How can you possibly tell us that hydrogen bonds are not needed for DNA replication?’,” says Kool. “That was the centre of the helix. And people were so fixated on hydrogen bonds that it was hard to even conceive of alternatives.” Instead of forming hydrogen bonds — a property normally associated with hydrophilic, or water-loving, molecules — F and other shape-mimicking bases developed by Kool’s team were hydrophobic. Water repels them, which helps them to stabilize in the double helix. DNA is analogous to a stack of coins, says Kool, and staying in the stack shields an unnatural base from water.
Floyd Romesberg, a chemical biologist at the Scripps Research Institute, has expanded the repertoire of hydrophobic bases. Starting with molecules such as benzene and naphthalene, his team built “every imaginable derivative”, he says. “It drove us very much away from anything that looked like a natural base pair at all.” But while testing steps in the replication process, the researchers found two contradictory requirements. A crucial position in the base had to be hydrophobic for enzymes to insert the base into DNA, yet it also had to accept hydrogen bonds if enzymes were to continue with copying the strand.
Romesberg’s team screened 3,600 combinations of 60 bases for the pair that was copied the most efficiently and accurately8. The two that won, MMO2 and SICS, “walk a thin line” between being hydrophobic and hydrophilic at the key position, Romesberg says.
A major challenge remained, however: researchers had to show that DNA would retain the unnatural base pairs while billions of copies are made. If enzymes pair unnatural with natural bases too often, the new letters could eventually disappear.
Base jumping
Ichiro Hirao, a chemist at the RIKEN Systems and Structural Biology Center in Yokohama, Japan, had been intrigued by the idea of creating unnatural bases ever since reading James Watson’s 1968 book The Double Helix as a teenager. Hirao and his colleagues found that they could reduce mispairing by designing shapes that fit awkwardly with natural bases, and by adding negatively charged or electron-rich chemical groups that repel the natural bases’ corresponding parts. In 2011, Hirao’s team reported that DNA containing an unnatural hydrophobic base pair, called Ds and Diol1-Px, could be copied with 99.77–99.92% fidelity per replication9. The same year, Benner and his colleagues showed that another unnatural base pair — P and Z, which join using hydrogen bonds — achieved fidelity of 99.8% per replication10. And in July, Romesberg’s team reported rates of 99.66–99.99% for optimized versions of his bases, called NaM and 5SICS (ref. 11), overlapping with the sloppiest rate for natural DNA. “Our best case is now approaching nature’s worst case,” says Romesberg.
Unnatural bases still have a lot to prove, however. Researchers haven’t shown that polymerases can copy more than four of the paired bases in a row10. The polymerase is “the hard nut to crack”, says Benner. And the solution may be to re-engineer it, too.
“If you were a chemist setting out to design this thing, you wouldn’t do it this way at all.”
Philipp Holliger, a chemical biologist at the Medical Research Council Laboratory of Molecular Biology in Cambridge, UK, and his colleagues demonstrated this approach earlier this year — using nucleic acids called XNA, in which the sugars normally present in DNA or RNA had been replaced by other ring structures12. The team generated billions of mutants of a natural polymerase and let them evolve by putting selective pressure on them to convert DNA to XNA (see Nature http://doi.org/jrh; 2012). The researchers then compared the most effective mutants to identify the best one. The polymerase is shaped roughly like a hand, and it turns out that the ‘thumb’ was the key region that needed to change, says Holliger. This region makes contact with the DNA as it exits the enzyme and might act as a final checkpoint to ensure correct synthesis. The team also engineered an enzyme that could convert XNA back into DNA.
Much of the tinkering so far has been done in vitro, but researchers hope to show that organisms can read and process the information. Perhaps the closest they have come to incorporating unnatural bases into a living system is an engineered bacterium reported last year13 by Philippe Marlière, co-founder of the microbial fluidics company Heurisko in Newark, Delaware. He and his team replaced most of the organism’s T bases with chlorouracil, a form of the RNA base uracil in which a hydrogen atom is replaced with chlorine. The team developed an automated system to introduce the base gradually to a strain of Escherichia coli that couldn’t make thymine on its own. After about five months, some of the bacteria couldn’t survive without chlorouracil and they had expunged roughly 90% of the thymine from their genomes.
Benner, Romesberg and Hirao are also working to coax cells to accept their base pairs. But even if the cells accept the pairs, they might have trouble carrying out processes such as recombination — a highly orchestrated reshuffling of genetic material. “It’s not just a matter of getting these darn things in,” says Andrew Ellington, a biochemist at the University of Texas at Austin and a former graduate student of Benner. “I think this is going to be a modestly Herculean task from here on out.”
Just how far researchers will get is unclear. Marlière’s team aims to replace all four natural bases with unnatural ones. But Romesberg says that developing an organism with only hydrophobic bases will be close to impossible, because cells contain too many components that have adapted to work with natural bases. As for combining an unnatural backbone and unnatural bases in one organism, “our theory is not good enough for us to go in and do both at the same time”, says Benner.
Even if unnatural base pairs don’t yet function in cells, they can still be put to practical use. Siemens Healthcare Diagnostics in Tarrytown, New York, and Luminex in Austin, Texas, already use Benner’s iso-C and iso-G pair to improve detection and monitoring of viral infections. Siemens, for example, uses a series of linked DNA sequences that bind to HIV-1 RNA in a patient’s blood sample. Inserting unnatural bases into some of the sequences discourages the sequences from binding to random DNA sequences in the sample and makes the HIV-1 RNA easier to detect at low levels.
DNA and RNA molecules can also catalyse reactions and be used as drugs. Developers can improve the performance of a sequence by attaching chemical groups to the bases, and unnatural bases make it easier to target a specific site in a sequence rather than saturating every C or G. Romesberg’s team has added ‘linker’ groups to unnatural bases in DNA that allow precise attachment of a variety of molecules. The team is now trying to engineer sequences that will catalyse reactions more efficiently than their natural counterparts.
Hirao says that his team has generated DNA sequences containing the Ds base that bind much better than natural sequences to interferon-?, an immune-system protein, and to vascular endothelial growth factor (VEGF), a therapeutic target in cancer and eye disease.
Practical applications aside, researchers are still driven by what Kool calls the “science-fiction appeal” of designing or even improving on living systems. Earth’s early life forms may have settled on their genetic alphabet simply because they were constrained by the chemicals available. Adenine, for example, is easy to make from hydrogen cyanide, which was probably present when life first emerged. Once organisms had a working set of bases, perhaps they got locked into that system. “If you start dabbling too much with your fundamental biochemistry, you’re going to get eaten,” says Benner. Although RNA — generally thought to have preceded DNA — might not be the best possible solution for supporting life, it might be the best solution that could have emerged on prebiotic Earth, Benner suggests.
So if nucleic acids arose independently on another planet, would they have the same bases? Benner thinks not, unless the organisms were subjected to the same constraints. Some universal rules might apply, however. For example, Benner says that backbones with repeating charges — which initially seemed to him like a liability — actually discourage folding and ensure that strands with different base sequences behave similarly during processes such as replication. Although some researchers have had success with alternative backbones, many attempts have resulted in molecules that are too stiff or too loose to form a helix. “I think there is a limit to the chemical variation that can be introduced,” says Holliger (see Nature 483, 528–530; 2012).
But that isn’t going to stop researchers from pushing the limits. “Why is the chemistry of living things the way it is? Is it because it’s the only possible answer?” asks Kool. “I believe the answer to that question is no. And the only way to prove it conclusively is to do it.”
- Nature
- 491,
- 516–518
- ()
- doi:10.1038/491516a
References
- Richert, C., Roughton, A. L. & Benner, S. A. J. Am. Chem. Soc. 118, 4518–4531 (1996).
- Switzer, C., Moroney, S. E. & Benner, S. A. J. Am. Chem. Soc. 111, 8322–8323 (1989).
- Piccirilli, J. A. et al. Nature 343, 33–37 (1990).
- Bain, J. D., Switzer, C., Chamberlin, A. R. & Benner, S. A. Nature 356, 537–539 (1992).
- Schweitzer, B. A. & Kool, E. T. J. Am. Chem. Soc. 117, 1863–1872 (1995).
- Moran, S., Ren, R. X.-F., Rumney, S. & Kool, E. T. J. Am. Chem. Soc. 119, 2056–2057 (1997).
- Moran, S., Ren, R. X.-F. & Kool, E. T. Proc. Natl Acad. Sci. USA 94, 10506–10511 (1997).
- Leconte, A. M. et al. J. Am. Chem. Soc. 130, 2336–2343 (2008).
- Yamashige, R. et al. Nucl. Acids Res. 40, 2793–2806 (2012).
- Yang, Z., Chen, F., Alvarado, J. B. & Benner, S. A. J. Am. Chem. Soc. 133, 15105–15112 (2011).
- Malyshev, D. A. et al. Proc. Natl Acad. Sci. USA 109, 12005–12010 (2012).
- Pinheiro, V. B. et al. Science 336, 341–344 (2012).
- Marlière, P. et al. Angew. Chem. Int. Edn 50, 7109–7114 (2011).